Abstract
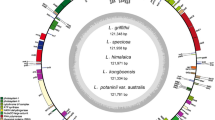
Species of the Pinus genus provide a classical model for studying hybrid speciation. Although studies on two narrowly distributed species (Pinus funebris and P. takahasii) concluded that they originated from two widespread species (P. sylvestris and P. densiflora) via hybrid speciation, the conclusion was based on a low number of informative restriction sites. In this study, we analyzed the sequences of four Pinus chloroplast (cp) genomes (P. sylvestris, P. densiflora, P. funebris and P. takahasii) to clarify whether hybrid speciation was involved. The complete cp-genomes of Pinus species ranged in size from 119,865 to 119,890 bp, similar to other Pinus species. Phylogenetic results based on the whole cp-genomes showed P. sylvestris clustered with P. funebris and P. takahasii, which suggested that P. sylvestris was the paternal parent in hybridization events. In an analysis of simple sequence repeats (SSRs), we detected a total of 69 SSRs repeats among the four Pinus cp-genomes; most were A or T bases. In addition, we identified divergent hotspot regions among the four Pinus cp-genomes (trnE-clpP, cemA-ycf4, petD-rpoA, psbD-trnT, and trnN-chlL), in P. sylvestris (psbD-trnT, trnN-chlL, psbB and rps8) and in P. densiflora (trnE-clpP, petD-rpoA, ycf3 intron, psbD-trnT, and trnN-chlL). The genome information found in this study provides new insights into hybrid speciation in Pinus and contributes to a better understanding of the phylogenetic relationships within the Pinus genus.






Similar content being viewed by others
Data availability
The complete chloroplast genome sequence data of the four Pinus species have been submitted to GenBank of NCBI: P. densiflora MT786135, P. sylvestris MT796488, P. takahasii MT786134, P. funebris MT793600. We released the raw data to the National Genomics Data Center (NGDC). The GSA DATA and submission is: CRA003870. The data will be available publicly upon publication.
References
Asaf S, Khan AL, Khan MA, Shahzad R, Kang SM, Al-Harrasi A, Al-Rawahi A, Lee IJ (2018) Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS ONE 13:e0192966
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS (2009) MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Byrne M, Hankinson M (2012) Testing the variability of chloroplast sequences for plant phylogeography. Aust J Bot 60:569–574
Chen Y, Hu N, Wu H (2019) Analyzing and characterizing the chloroplast genome of Salix wilsonii. Biomed Res Int 2019:1–14
Chen Y, Ye W, Zhang Y, Xu Y (2015) High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res 43:7762–7768
Curci PL, Paola DD, Danzi D, Vendramin GG, Sonnante G (2015) Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS ONE 10:e0120589
Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17:1–29
Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res 45:e18–e18
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf material. Photochem Bull 19:11–15
Fukuda T, Ashizawa H, Suzuki R, Ochiai T, Nakamura T, Kanno A, Kameya T, Yokoyama J (2005) Molecular phylogeny of the genus Asparagus (Asparagaceae) inferred from plastid petB intron and petD–rpoA intergenic spacer sequences. Plant Species Biol 20:121–132
Gao LZ, Liu YL, Zhang D, Li W, Gao J, Liu Y, Li K, Shi C, Zhao Y, Zhao YJ, Jiao JY, Mao SY, Gao CW, Eichler EE (2019) Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun Biol 2:1–13
Gault CM, Kremling KA, Buckler ES (2018) Tripsacum de novo transcriptome assemblies reveal parallel gene evolution with maize after ancient polyploidy. Plant Genome
Gernandt DS, López GG, García SO, Liston A (2005) Phylogeny and classification of Pinus. Taxon 54:29–42
Gross BL, Rieseberg LH (2005) The ecological genetics of homoploid hybrid speciation. J Hered 96:241–252
He Y, Xiao H, Deng C, Xiong L, Yang J, Peng C (2016) The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int J Mol Sci 17(6):820
Hegarty MJ, Hiscock SJ (2008) Genomic clues to the evolutionary success of polyploid plants. Curr Biol 18:R435–R444
Jeong YM, Chung WH, Mun JH, Kim N, Yu HJ (2014) De novo assembly and characterization of the complete chloroplast genome of radish (Raphanus sativus L.). Gene 551:39–48
Julca I, Marcet-houben M, Vargas P, Gabaldón T (2018) Phylogenomics of the olive tree (Olea europaea) reveals the relative contribution of ancient allo- and autopolyploidization events. BMC Biol 16:1–15
Kang HI, Lee HO, Lee IH, Kim IS, Lee SW, Yang TJ, Shim D (2019) Complete chloroplast genome of Pinus densiflora Siebold & Zucc and comparative analysis with five pine trees. Forests 10:600
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kim S, Li G, Han SH, Chang H, Kim HJ, Son Y (2017) Differential effects of coarse woody debris on microbial and soil properties in Pinus densiflora Sieb. ET Zucc Forests Forests 8:292
Kong H, Liu W, Yao G, Gong W (2017) A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): a traditional herbal medicinal genus. PeerJ 5:e4018
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X (2012) CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom 13:715
Liu X, Li Y, Yang H, Zhou B (2018) Chloroplast genome of the folk medicine and vegetable plant Talinum paniculatum (Jacq) Gaertn: gene organization, comparative and phylogenetic analysis. Molecules 23:857
Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52:267–274
Minami M, Nishio K, Ajioka Y, Kyushima H, Shigeki K, Kinjo K, Yamada K, Satoh K, Nagai M, Sakurai Y (2009) Identification of Curcuma plants and curcumin content level by DNA polymorphisms in the trnS-trnfM intergenic spacer in chloroplast DNA. J Nat Med 63:75–79
Ni L, Zhao Z, Xu H, Chen S, Dorje G (2017a) Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr Genet 63:241–252
Ni Z, Ye Y, Bai T, Xu M, Xu LA (2017b) Complete chloroplast genome of Pinus massoniana (Pinaceae): gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 22:1528
Nie X, Deng P, Feng K, Liu P, Du X, You FM, Song WN (2014) Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol Biol Report 32:828–840
Park I, Kim WJ, Yang S, Yeo SM, Li H, Moon BC (2017) The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS ONE 12:e0184257
Peters D, Qiu K, Liang P (2011) Faster short DNA sequence alignment with parallel BWA. AIP Conf Proc 1368:131–134
Ran JH, Shen TT, Liu WJ, Wang PP, Wang XQ (2015) Mitochondrial introgression and complex biogeographic history of the genus Picea. Mol Phylogenet Evol 93:63–76
Rassi FG, Abra ML, Cienza AS, Mazio SI (2002) Chloroplast SSR markers to assess DNA diversity in wild and cultivated grapevines. Vitis 41:157–158
Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom 8:174
Ren GP, Abbott RJ, Zhou YF, Zhang LR, Peng YL, Liu JQ (2012) Genetic divergence, range expansion and possible homoploid hybrid speciation among pine species in Northeast China. Heredity (edinb) 108:552–562
Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Mei WB, Thomas PI, Gardner MF, Ennos RA, Hollingsworth PM (2015) Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Resour 15:1067–1078
Savolainen O, Lascoux M, Merilä J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14:807–820
Seixas FA, Boursot P, Melo-Ferreira J (2018) The genomic impact of historical hybridization with massive mitochondrial DNA introgression. Genome Biol 19:1–20
Staton SE, Ungerer MC, Moore RC (2009) The genomic organization of Ty3/gypsy-like retrotransposons in Helianthus (Asteraceae) homoploid hybrid species. Am J Bot 96:1646–1655
Tang D, Wei F, Cai Z, Wei Y, Khan A, Miao J, Wei K (2021) Analysis of codon usage bias and evolution in the chloroplast genome of Mesona chinensis Benth. Dev Genes Evol 231:1–9
Team RC (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Tyler T, Jönsson J (2013) Patterns of plastid and nuclear variation among apomictic polyploids of Hieracium: evolutionary processes and taxonomic implications. Ann Bot 111:591–609
Vallejo-Marín M, Hiscock SJ (2016) Hybridization and hybrid speciation under global change. New Phytol 211:1170–1187
Watano Y, Kanai A, Tani N (2004) Genetic structure of hybrid zones between Pinus pumila and P. parviflora var. pentaphylla (Pinaceae) revealed by molecular hybrid index analysis. Am J Bot 91:65–72
Weng M, Ruhlman TA, Jansen RK (2017) Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol 214:842–851
Wicker T, Gundlach H, Spannagl M, Uauy C, Borrill P, Ramírez-González RH, Oliveira RD, IWGSC, Mayer KFX, Paux E, Choulet F (2018) Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol 19:103
Wu X, Fan Y, Li L, Liu Y (2020) The influence of soil drought stress on the leaf transcriptome of faba bean (Vicia faba L.) in the Qinghai-Tibet Plateau. 3 Biotech 10:381
Xue Y, Shao LP, Wang ZH (1990) A study on Xingkai Lake pine gall rust. J Northeast for Univ 1:65–71
Yakimowski SB, Rieseberg LH (2014) The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. Am J Bot 101:1247–1258
Yu T, Huang BH, Zhang Y, Liao PC, Li JQ (2020) Chloroplast genome of an extremely endangered conifer Thuja sutchuenensis Franch.: gene organization, comparative and phylogenetic analysis. Physiol Mol Biol Plants 26:409–418
Zaborowska J, Łabiszak B, Wachowiak W (2020) Population history of European mountain pines Pinus mugo and Pinus uncinata revealed by mitochondrial DNA markers. J Syst Evol 58:474–486
Zeb U, Dong WL, Zhang TT, Wang RN, Shahzad K, Ma XF, Li ZH (2020) Comparative plastid genomics of Pinus species: Insights into sequence variations and phylogenetic relationships. J Syst Evol 58:118–132
Zhao J, Xu Y, Xi L, Yang J, Chen H, Zhang J (2018) Characterization of the chloroplast genome sequence of acer miaotaiense: comparative and phylogenetic analyses. Molecules 23:1740
Zonneveld BJM (2012) Conifer genome sizes of 172 species, covering 64 of 67 genera, range from 8 to 72 picogram. Nord J Bot 30:490–502
Funding
This research was funded by Kunyu Mountain National Nature Reserve Administration.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This research was funded by Kunyu Mountain National Nature Reserve Administration.
The online version is available at http://www.springerlink.com
Corresponding editor: Yanbo Hu.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, T., Jia, Z., Dayananda, B. et al. Analysis of the chloroplast genomes of four Pinus species in Northeast China: Insights into hybrid speciation and identification of DNA molecular markers. J. For. Res. 33, 1881–1890 (2022). https://doi.org/10.1007/s11676-021-01432-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-021-01432-7